Page 41 - Demo
P. 41
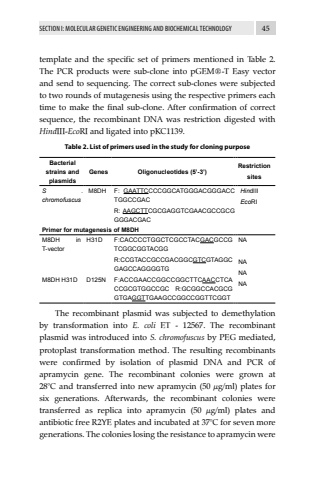
SECTION I: MOLECULAR GENETIC ENGINEERING AND BIOCHEMICAL TECHNOLOGY 45template and the specific set of primers mentioned in Table 2. The PCR products were sub-clone into pGEM%u00ae-T Easy vector and send to sequencing. The correct sub-clones were subjected to two rounds of mutagenesis using the respective primers each time to make the final sub-clone. After confirmation of correct sequence, the recombinant DNA was restriction digested with HindIII-EcoRI and ligated into pKC1139. Table 2. List of primers used in the study for cloning purposeBacterial strains and plasmidsGenes Oligonucleotides (5%u2019-3%u2019)RestrictionsitesS . chromofuscusM8DH F: GAATTCCCCGGCATGGGACGGGACCTGGCCGACR: AAGCTTCGCGAGGTCGAACGCCGCGGGGACGACHindIIIEcoRIPrimer for mutagenesis of M8DHM8DH in T-vectorH31D F:CACCCCTGGCTCGCCTACGACGCCGTCGGCGGTACGGR:CCGTACCGCCGACGGCGTCGTAGGCGAGCCAGGGGTGNANANANA M8DH H31D D125N F:ACCGAACCGGCCGGCTTCAACCTCACCGCGTGGCCGC R:GCGGCCACGCGGTGAGGTTGAAGCCGGCCGGTTCGGTThe recombinant plasmid was subjected to demethylation by transformation into E. coli ET - 12567. The recombinant plasmid was introduced into S. chromofuscus by PEG mediated, protoplast transformation method. The resulting recombinants were confirmed by isolation of plasmid DNA and PCR of apramycin gene. The recombinant colonies were grown at 28%u00b0C and transferred into new apramycin (50 %u00b5g/ml) plates for six generations. Afterwards, the recombinant colonies were transferred as replica into apramycin (50 %u00b5g/ml) plates and antibiotic free R2YE plates and incubated at 37%u00b0C for seven more generations. The colonies losing the resistance to apramycin were